Permanent changes in the DNA sequence, called mutations, can have serious consequences for cells and are repaired through mismatch repair, chemical reversal, excision repair, and double-stranded break repair.
Some errors are not corrected during replication, but are instead corrected immediately after replication is completed; this type of repair is known as mismatch repair. Mismatch repair can also correct small insertions and deletions that happen when the DNA polymerase slips on a template. If a mismatch error occurs, a protein complex recognizes and binds to the mispaired base. A second complex cuts the DNA near the mismatch, and more enzymes chop out the incorrect nucleotide and a surrounding patch of DNA. A DNA polymerase then replaces the missing section with correct nucleotides, and an enzyme called DNA ligase seals the gap.

Damage to DNA can occur at any point, not just during replication. Some sources of damage that can induce mutations in the DNA sequence include UV light, chemicals, and X-rays. These types of damage can be repaired in a few different ways.
In some cases, a cell can fix DNA damage simply by reversing the chemical reaction that caused it. Sometimes, DNA damage can refer to an extra group of atoms getting attached to DNA through a chemical reaction. For example, an unwanted methyl group can attach to the nucleotide guanine, as shown below; this chemical reaction can be reversed by a specific enzyme.

Another mechanism, called nucleotide excision repair, can remove and replace damaged nucleotides that warp the DNA helix. For example, UV radiation can make cytosine and thymine bases react, forming bonds that distort the double helix and cause errors in DNA replication. The most common type of linkage, a thymine dimer, consists of two thymine bases that react with each other. In nucleotide excision repair, a helicase separates the two DNA strands around the damaged nucleotide(s), which are then removed along with a surrounding patch of DNA, as shown below. A DNA polymerase replaces the missing DNA, and a DNA ligase seals the backbone.

Base excision repair is a mechanism used to detect and remove certain types of damaged bases. A group of enzymes called glycosylases play a key role in base excision repair. Each glycosylase detects and removes a specific kind of damaged base. For example, a chemical reaction called deamination can convert a cytosine base into uracil, a base typically found only in RNA, as shown below. Glycosylase can remove the incorrect uracil base, DNA polymerase can fill in the gap, and DNA ligase will again seal the backbone.

Finally, the most serious type of DNA damage is a double-stranded DNA break, which can cause large regions of chromosomes to be lost if the break isn’t repaired. There are two pathways involved in double-stranded break repair: non-homologous end joining and homologous recombination.
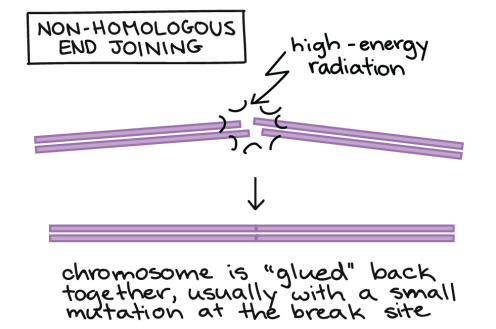
In non-homologous end joining, the two broken ends of the chromosome are glued back together. This repair mechanism is “messy” and typically involves the loss, or sometimes addition, of a few nucleotides at the cut site. This introduces a small mutation, but is more beneficial than a region of chromosome being lost.
In homologous recombination, information from the homologous chromosome that matches the damaged one (or from a sister chromatid, if the DNA has been copied) is used to repair the break. In this process, the two homologous chromosomes come together, and the undamaged region of the homologue or chromatid is used as a template to replace the damaged region of the broken chromosome. Homologous recombination is “cleaner” than non-homologous end joining and does not usually cause mutations.

Key Points
• When replication mistakes are not corrected, they may result in mutations.
• Mutations can also arise from external sources, like UV radiation.
• Mutations can sometimes be benign, but can also have serious consequences, like cancer.
• Mismatch repair occurs immediately after DNA replication. Enzymes involved in this type of repair recognize incorrectly incorporated bases, remove them from DNA, and replace them with the correct bases.
• In nucleotide excision repair, one or more damaged nucleotides and a small surrounding DNA region are removed and replaced.
• In base excision repair, a single incorrect base is removed and filled in.
• Double-stranded DNA breaks are the most serious type of DNA damage.
• Double-stranded DNA breaks can be repaired by non-homologous end joining, which introduces a small mutation, or homologous recombination, which uses a homologous chromosome as a template for the repair.
Key Terms
Mutation: Errors in the DNA sequence that arise during replication or due to environmental factors like UV light and chemical reactions.
Mismatch repair: A system for recognizing and repairing some forms of DNA damage and erroneous insertion, deletion, or misincorporation of bases that can arise during DNA replication and recombination
Chemical reversal: A DNA repair mechanism based on the reversal of chemical reactions that induce DNA damage.
Nucleotide excision repair: A DNA repair mechanism that corrects damage done by UV radiation, including thymine dimers, that cause bulky distortions in the DNA helix.
Base excision repair: A DNA repair mechanism that detects and removes damaged nucleotide bases.
Double-stranded break repair: A DNA repair mechanism that can correct simultaneous breaks in both strands of a DNA molecule through either non-homologous end joining or homologous recombination.
DNA polymerase: An enzyme that adds successive nucleotides to a growing DNA strand during DNA replication; this protein also has proofreading and exonuclease activity to ensure the correct nucleotides are added.
DNA ligase: An enzyme that joins nucleotides by catalyzing the formation of a phosphodiester linkage.
Thymine dimer: A type of DNA damage resulting from the chemical linkage of two thymine bases with each other.
Glycosylases: Enzymes that mediate base excisions repair.
Non-homologous end joining: A repair mechanism that involves connecting the two broken ends of a chromosome together; usually involves the loss or addition of a few nucleotides and therefore the introduction of a mutation.
Homologous recombination: A repair mechanism that uses a homologous chromosome or sister chromatid as the template for repair of a double-stranded DNA break.









